Combating SARS-CoV-2 by induced error catastrophy?
Research group of Prof. Dr. Chris Meier and postdoc Dr. Johanna Huchting, Universität Hamburg, in joint project with Dr. Bruno Canard and the Viral Replication group at the University of Aix-Marseille in France.
Publication information:
Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis
Ashleigh Shannon1, Barbara Selisko1, Nhung-Thi-Tuyet Le1, Johanna Huchting2, Franck Touret3, Géraldine Piorkowski3, Véronique Fattorini1, François Ferron1, Etienne Decroly1, Chris Meier2, Bruno Coutard3, Olve Peersen4* & Bruno Canard1*
Nature Communications DOI: 10.1038/s41467-020-18463-z 17th September 2020
1. Summary
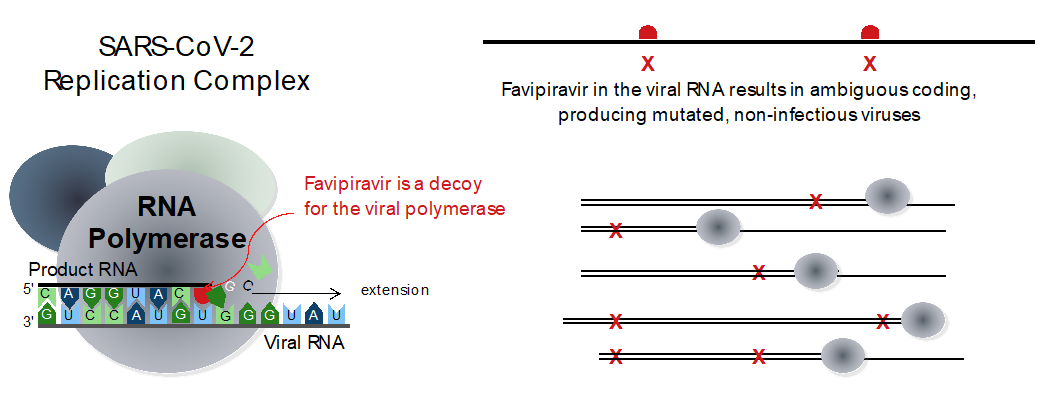
SARS-CoV-2, the virus responsible for the COVID-19 pandemic, is a coronavirus whose genetic material is reproduced during infection by a viral polymerase enzyme. The results from this research show the coronavirus polymerase is extraordinary in being ten times faster than other viral polymerases, but also much less faithful and it produces errors at a very high rate. Many of these errors are repaired by a viral error correcting enzyme, but by using the antiviral drug Favipiravir (Avigan) the researchers have managed to make the virus accumulate too many mutations, overloading the error correcting machinery and rendering the viral genome non-functional.
2. Thumbnail Image
3. Research Description
The SARS-CoV2 virus, like its SARS-CoV-1 twin from 2003, is part of the Coronavirus family with seven members that infect humans. Of these, only SARS-CoV-1 and 2 and MERS-CoV are highly pathogenic, and SARS-CoV-2 is the most contagious, as can be seen with the difficulties trying curb the current pandemic that is approaching one million deaths and more than 20 million infections.
The coronaviruses use RNA as their genetic material and they have genomes that are 3-4 times larger than the "classic" RNA viruses (Dengue, Flu, Ebola, Hepatitis C, Polio). The coronavirus genome encodes for many viral proteins, some of which allow the viruses to evade the cell's immune response, while others allows the virus to grow by replicating their genomes. A central protein of the replication process is the viral RNA polymerase, an enzyme that has no functional equivalent in human cells and is therefore a very interesting target for antiviral treatments. For some viruses, including HIV, hepatitis C, herpes viruses and influenza virus, there exist drugs that target the viral polymerase in the form of nucleoside analogues. These compounds can be incorporated into the viral RNA by the polymerase in place of the four natural genetic bases, and doing so stops further synthesis of the genome and kills the virus. But coronaviruses are unique in that they have an added exonuclease enzyme that is capable of repairing the damage caused by many of these drugs, reducing their antiviral utility. Remdesivir is at least partially resistant to this repair, contributing to its success against SARS-CoV-2, but this drug must be administered intravenously and its dosage is delicate.
Researchers from AFMB-CNRS, the University of Aix-Marseille, the University of Hamburg, and Colorado State University report the first detailed kinetic and mechanistic analysis of the CoV polymerase enzyme, using the anti-influenza drug Avigan (i.e. Favipiravir or T-705). Avigan is already on the market in Japan and in clinical trials for several months in Russia, China, Iran, India, Japan and the United States. It does not stop the synthesis of viral RNA, but rather allows the synthesis of genetically altered genomes, which pushes the virus towards an evolutionary and functional impasse and ultimately produces an antiviral effect via a mechanism called “error catastrophe.”
The researchers discovered two quite remarkable properties of the SARS-CoV-2 RNA polymerase: in order to synthesize these very long genomes, the RNA polymerase has evolved an extraordinarily high catalytic efficiency and replicates the RNA at very high speeds that is about ten times faster than similar RNA virus polymerases. At the same time, and under normal conditions of infection, the data show the polymerase has sacrificed its capacity for genetically faithful synthesis, which is possible because the coronaviruses have added an error-repairing exonuclease enzyme that can help restore a faithful copy of the genome.
From theses results, a new concept emerges in the design of anti-coronavirus drugs using nucleoside analogue compounds that look similar to the normal genetic bases: Perhaps it is better to take advantage of the extraordinary low fidelity of the coronavirus polymerase to deceive the enzyme by making it synthesize a lot of errors, so many that it overloads the error correcting mechanism and allowing the virus "over-mutate" until it dies. This may be more efficient than trying to stop both viral synthesis and repair steps at the same time.
The extraordinary fast and low fidelity coronavirus polymerase also raises many questions about the evolution and size of the RNA viruses. Could a fast and error-correcting replication complex have allowed for the increased genome size and associated capacity for gene acquisition in these viruses? Striking similarities with the genomes of small DNA viruses suggests that coronaviruses could represent a particularly interesting study model for understanding how living organisms switched, long ago, from using RNA to using DNA for their genomes.
