Venom Proteomics
In collaboration with international partners we initiated a project whose long-term goal is the detailed analysis and comparison of selected venom proteomes, using biochemical and biophysical methods. Venoms represent the critical innovation in ophidian evolution that allowed advanced snakes to transition from a mechanical (constriction) to a chemical (venom) means of subduing and digesting large prey, and as such, venom proteins have multiple functions including immobilizing, paralyzing, killing and digesting prey.
Snake venom is a mixture of low and high molecular mass polypeptides, metal ions, carbohydrates, nucleosides, amines and a very small amount of lipids, and free amino acids. Proteins predominate and represent 90 – 95% of the venom dry weight. Snake bite is a serious medical problem worldwide and approx. 2.5 million humans are affected annually and approx. 100 000 die. Snakebites affects breathing, the homeostatic system and tissue repair, and cause neurotoxicity, myonecrosis, intravascular haemolysis, haemorrhage, coagulopathy, renal failure and increase capillary permeability. A detailed knowledge of the toxin composition of the venoms is very important for an in-depth understanding of the envenomation mechanisms and treatment of patients suffering of the snakebite consequences, as well as phylogenetic analysis of the toxins and biology and evolution of the snake is today most important to obtain most insights about evolutionary aspects. Within a systematic approach a research topic is focused to analyze first the protein components by 2-D electrophoresis, followed by in-gel trypsinolysis. The cleaved peptides are eluted and subjected to LC/MS/MS analysis. So far we have determined the entire venom proteomes of several Viperidae species in collaboration with scientists at the Sao Paolo State University, Brazil, the All India Institute of Medical Sciences and the Academy of Sciences Bulgaria.
Snake venom is a mixture of low and high molecular mass polypeptides, metal ions, carbohydrates, nucleosides, amines and a very small amount of lipids, and free amino acids. Proteins predominate and represent 90 – 95% of the venom dry weight. Snake bite is a serious medical problem worldwide and approx. 2.5 million humans are affected annually and approx. 100 000 die. Snakebites affects breathing, the homeostatic system and tissue repair, and cause neurotoxicity, myonecrosis, intravascular haemolysis, haemorrhage, coagulopathy, renal failure and increase capillary permeability. A detailed knowledge of the toxin composition of the venoms is very important for an in-depth understanding of the envenomation mechanisms and treatment of patients suffering of the snakebite consequences, as well as phylogenetic analysis of the toxins and biology and evolution of the snake is today most important to obtain most insights about evolutionary aspects. Within a systematic approach a research topic is focused to analyze first the protein components by 2-D electrophoresis, followed by in-gel trypsinolysis. The cleaved peptides are eluted and subjected to LC/MS/MS analysis. So far we have determined the entire venom proteomes of several Viperidae species in collaboration with scientists at the Sao Paolo State University, Brazil, the All India Institute of Medical Sciences and the Academy of Sciences Bulgaria.
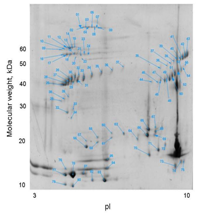 2-D gel electrophoresis of the venom from the Siamese Russell’s viper (Daboia russelli siamensis). |
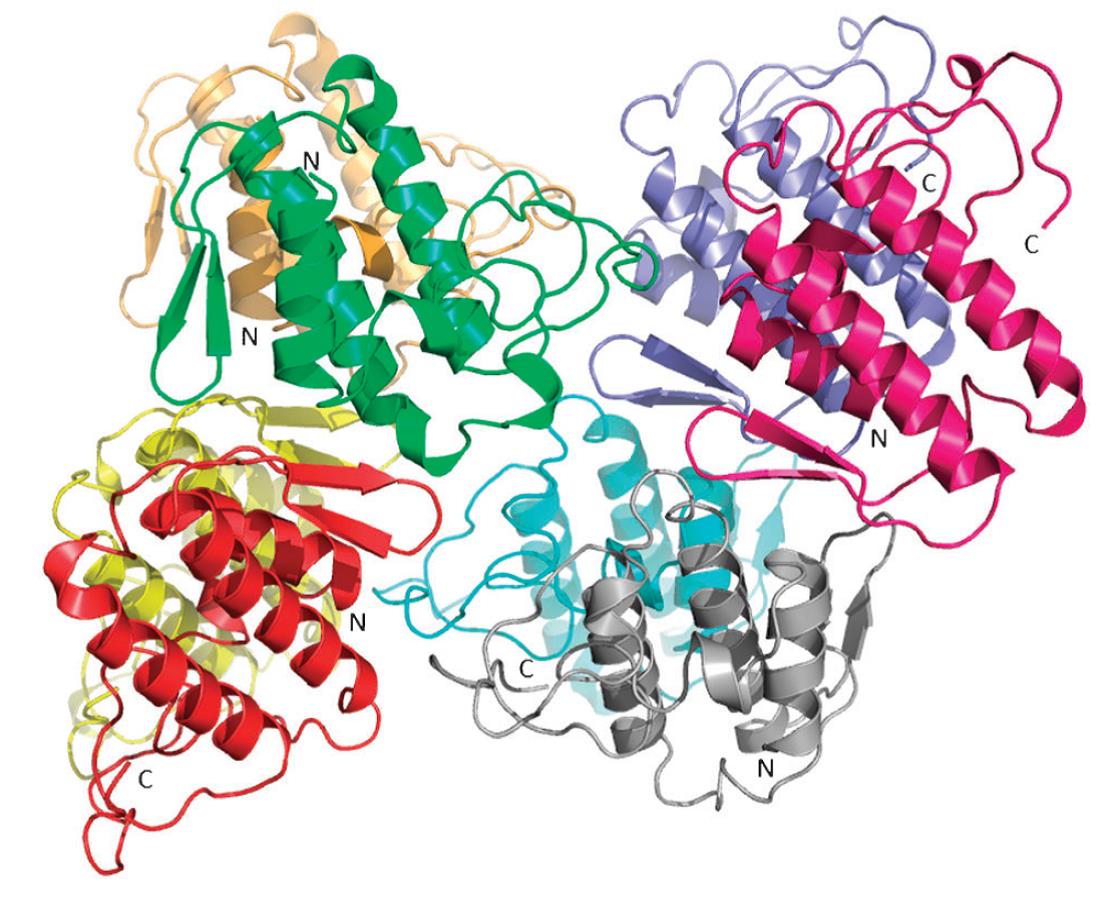 3D structure of 4 PlA2 dimers in one asymmetric unit of a Vipera ammodytes meridionalis PLA2-like myotoxin crystal structure. |
For structure-function-analysis we further isolate and characterize novel venom proteins affecting blood coagulation and clotting, tumor growth, neurotoxins, myotoxins, cardiotoxins, activators and inhibitors of inflammation, nerve growth factors, etc. For example the 3D structure of the neurotoxic PLA2 from the venom of Vipera ammodytes meridionalis in a complex with the powerful inhibitor elaidoylamide revealed a new mode of Asp49 PLA2 inhibition: the hydrocarbon chain of a fatty acid is bound simultaneously to the substrate binding channels of two PLA2 molecules. The structure is a suitable starting point to design new anti-inflammatory drugs, because PLA2 is involved in several inflammatory processes. One of our main goals of the future determination snake venom proteomes is the creation of a proteome based Venom Toxin Data Base, which will be of interest for physicians, toxycologists, pharmacologists, clinical laboratories and scientists worldwide.
Selected publications
- A. Ullah, R. Masood, I. Ali, K. Ullah, H. Ali, H. Akbar, Ch. Betzel: Thrombin-like enzymes from snake venom: Structural characterization and mechanism of action. Int. J. Biol. Macromol. 114, 788-811 (2018).
- R. Masood, K. Ullah, H. Ali, I. Ali, Ch. Betzel, A. Ullah: Spider's venom phospholipases D: A structural review. Int. J. Biol. Macromol. 107, 1054-1065 (2018).
- D.N. Georgieva, D. Hildebrand, R. Simas, M. A. Coronado, M. Kwiatkowski, H. Schlüter, R. K. Arni, P.J. Spencer, C. Betzel: Protein profile analysis of two Australian snake venoms by one-dimensional gel electrophoresis and MS/MS experiments. Current Medicinal Chemistry 24, 1892-1908 (2017)
- A. Munawar, D. Georgieva, Ch. Betzel, M. Trusch, H. Behnken, D. Hildebrand, M. Kwiatkowski, S. Harder, H. Schlüter, R. Arni, P. Spencer: Elapid Snake Venom Analyses Show the Specificity of the Peptide Composition at the Level of Genera Naja and Notechis. Toxins 6, 850-868 (2014).
- M. Coronado, A. Gabdulkhakov, D. Georgieva, B. Sankaran, M. Murakami, R. K. Arni, Ch. Betzel: Structure of the polypeptide crotamine from the Brazilian rattlesnake Crotalus durissus terrificus. Acta Crystallographica, Section D Biological Crystallography D69, 1958-1964 (2013).
- A. Munawar, M. Trusch, D. Georgieva, P. Spencer, V. Frochaux, S. Harder, R.K. Arni, D. Duhalov, N. Genov, H. Schlüter, Ch. Betzel: Venom peptide analysis of Vipera ammodytes meridionalis (Viperinae) and Bothrops jararacussu (Crotalinae) demonstrates subfamily-specificity of the peptidome in the family Viperidae. Mol. Biosyst. 7, 3298-307 (2011).
- D. Georgieva, J. Seifert, M. Öhler, M. von Bergen, P. Spencer, R.K. Arni, N. Genov, Ch. Betzel: Pseudechis australis venomics: adaptation for a defense against microbial pathogens and recruitment of body transferrin. J. Proteome Res. 10, 2440-64 (2011).
- D. Georgieva, M. Murakami, M. Perband, R.K. Arni, Ch. Betzel: The structure of a native l-amino acid oxidase, the major component of the Vipera ammodytes ammodytes venomic, reveals dynamic active site and quaternary structure stabilization by divalent ions. Mol Biosyst. 7, 379-84 (2011).
- E.A. Undheim, D. Georgieva, H.H. Thoen, J.A. Norman, J. Mork, Ch. Betzel, B.G. Fry: Venom on ice: first insights into Antarctic octopus venoms. Toxicon. 56, 897-913 (2010).
- M. Ohler, D. Georgieva, J. Seifert, M. von Bergen, R.K. Arni, N. Genov, Ch. Betzel: The venomics of Bothrops alternatus is a pool of acidic proteins with predominant hemorrhagic and coagulopathic activities. J. Proteome Res. 9, 2422-37 (2010).
- D. Georgieva, M. Ohler, J. Seifert, M. von Bergen, R.K. Arni, N. Genov, Ch. Betzel: Snake venomic of Crotalus durissus terrificus--correlation with pharmacological activities. J. Proteome Res. 9, 2302-16 (2010).
- D. Georgieva, R. Arni, Ch. Betzel: Proteome analysis of snake venom toxins: pharmacological insights. Expert Review of Proteomics 5, 787-797 (2008), Review.
