Phasing with triangles
Previous work: Protein structure determination with halogenated ligands
This project was started during the PhD thesis of TB. A new class of compounds was developed that can be used for heavy-atom derivatization of macromolecules to solve new protein structures with X-ray crystallography. The compounds combine heavy atoms for experimental phasing with functional groups for hydrogen bonding to biomolecules.
The magic triangle I3C contains three iodine atoms. The anomalous signal at in-house X-ray sources can be used for single-wavelength anomalous dispersion (SAD) experiments. The MAD triangle B3C has the same functional groups as I3C, but the three bromine atoms present allow for experimental phasing using multi-wavelength anomalous dispersion (MAD). In I3C and B3C, the heavy atoms form an equilateral triangle that can be easily identified in the heavy-atom substructure. The functional groups make the compounds sticky: several types of interaction through hydrogen bonds are observed in the crystal structures. Hydrogen bonds involve side chains but also the main chain of proteins.
I3C: The magic triangle
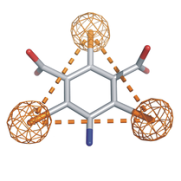
The magic triangle I3C
The triiodo benzene derivative 5-amino-2,4,6-triiodoisophthalic acid (I3C) can be used for heavy atom derivatisation of biological macromolecules.
Three functional groups for hydrogen bonding along with three iodine atoms render I3C a suitable agent for experimental phasing of macromolecules. The iodine atoms give rise to a high anomalous signal, even at in-house sources. Additionally, they form an equilateral triangle (with a side of 6.0 Å) which is easily recognised in the heavy atom substructure when this compound is used for heavy atom derivatisation in macromolecular phasing
Phasing with I3C
The compound I3C has been incorporated into protein crystals using cocrystallisation and soaking procedures. Jena Bioscience has developed the JBS Magic Triangle Kit that includes all required chemicals and an extensive user guide. I3C is also available directly from different chemical suppliers.
The iodine atoms present give rise to a strong anomalous signal that can be used for phasing. The triangle is readily identified in the heavy atom substructure and can point to a correct substructure solution. The two carboxylate groups and the amino group form hydrogen bonds to the main chain and to side chains of the protein. For further details on the experiment and phasing, please refer to:
|
Beck, T., Krasauskas, A., Gruene, T. & Sheldrick, G.M.: "A magic triangle for experimental phasing of macromolecules." Acta Crystallogr. Section D 2008, 64, 1179-1182. [pdf] |
I3C has also been used for phase determination with SIRAS methods:
| Sippel, K. H., Robbins, A. H., Reutzel, R., Domsic, J., Boehlein, S. K., Govindasamy, L., Agbandje-Mckenna, M., Rosser, C. J., Mckenna, R.: "Structure determination of the cancer-associated mycoplasma hyorhinis protein Mh-p37." Acta Crystallogr. Section D 2008, 64, 1172-1178. |
Incorporation of I3C
Usually I3C is incorporated by soaking a protein crystal with a solution of I3C. This soaking solution contains the same buffer and precipitant concentration as the crystallisation condition, but in addition a high concentration of I3C (e.g. 0.5 M). It is recommended to start with a high I3C concentration and a short soaking time (quick soak). Soaking times as short as 10 seconds proved to be sufficient for successful incorporation of I3C. If crystal degradation occurs at high concentrations, a lower concentration along with a longer soaking time or a gradient soak is recommended.
I3C may also be incorporated by means of cocrystallisation. I3C is added directly to the crystallisation drop at a lower concentration than used for soaking.
Data collection
Several structures have been solved with data obtained from in-house source (Cu Kalpha). If possible, data sets should have a high multiplicity to ensure that the anomalous signal is accurately determined. Structure solution can be carried out by SAD phasing. If you have an isomorphous data set from a native crystal, it is recommended to try SIRAS phasing to combine the isomorphous differences and the anomalous signal for phasing.
Restraint and model files for I3C
Once I3C has been incorporated into the protein crystal and data have been collected, it is either possible to refine the native data set (SIRAS) or to refine the derivative data set with I3C present in the crystal lattice (SAD). For the refinement of the derivative data set, restraints for I3C are required. Restraints for two popular refinement programs (refmac and SHELX) have been set up. There is also a model file available. This can be loaded into COOT, fitted in the density and merged with the protein PDB file to set up a file for refinement. All files have been derived from the crystal structure of I3C:
| Beck, T. & Sheldrick, G. M.: "5-Amino-2,4,6-triiodoisophthalic acid monohydrate." Acta Crystallogr. Section E 2008, 64, o1286. [pdf] |
In general, the use of a server/database is also an option for obtaining restraints and model files. However, not all required restraints (e.g. the planarity restraint for the carboxylate group) are usually included. Thus, great care has to be taken when using those generated restraints and model files.
For the I3C files, please contact TB. If you require any assistance with the refinement of I3C in your structure, please also contact TB.
References
"A magic triangle for experimental phasing of macromolecules." Acta Crystallogr. Section D 2008, 64, 1179-1182.
"5-Amino-2,4,6-triiodoisophthalic acid monohydrate." Acta Crystallogr. Section E 2008, 64, o1286.
B3C: The MAD triangle
At the 2009 CCP4 Study Weekend in Nottingham, the bromine derivative B3C was introduced. It can be used for multi-wavelength anomalous dispersion (MAD) experiments. It contains the same functional groups as I3C and therefore interacts also through hydrogen bonds with biological macromolecules.
If you would like to try MAD phasing, we actually recommend the MAD tetragon B4C. This compound is commercially available. In general we found that I3C binds better to proteins compared with B3C. A SAD dataset of an I3C-derivatized crystal, either collected in-house or at a long wavelength at the synchrotron, together with a native dataset (if available) can provide sufficient phase information to solve structures by SAD or SIRAS.
References
"The magic triangle goes MAD: experimental phasing with a bromine derivative." Acta Crystallogr. Section D 2010, 66, 374-380. open access [pdf]
"5-Amino-2,4,6-tribromoisophthalic acid: the MAD triangle for experimental phasing." Acta Crystallogr. Section C 2009, 65, o237-o239. [pdf]
B4C: The MAD tetragon
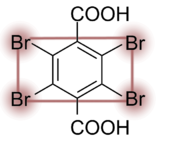
The MAD tetragon B4C
The latest compound is the MAD tetragon B4C. It was introduced at the ECM 26 Meeting in Darmstadt. B4C has two functional groups for interaction with biological macromolecules and four bromine atoms for experimental phasing via SAD or MAD.
B4C is commercially available: its chemical name is tetrabromoterephthalic acid. It can be ordered from any chemical supplier. The usage of B4C is similar to the I3C and B3C compounds.
In general we found that I3C usually binds better to proteins compared with B3C and B4C. A SAD dataset of an I3C-derivatized crystal, either collected in-house or at a long wavelength at the synchrotron, together with a native dataset (if available) can provide sufficient phase information to solve structures by SAD or SIRAS.
For further information or enquiries, please contact TB.
