Research
The ribosome and protein synthesis
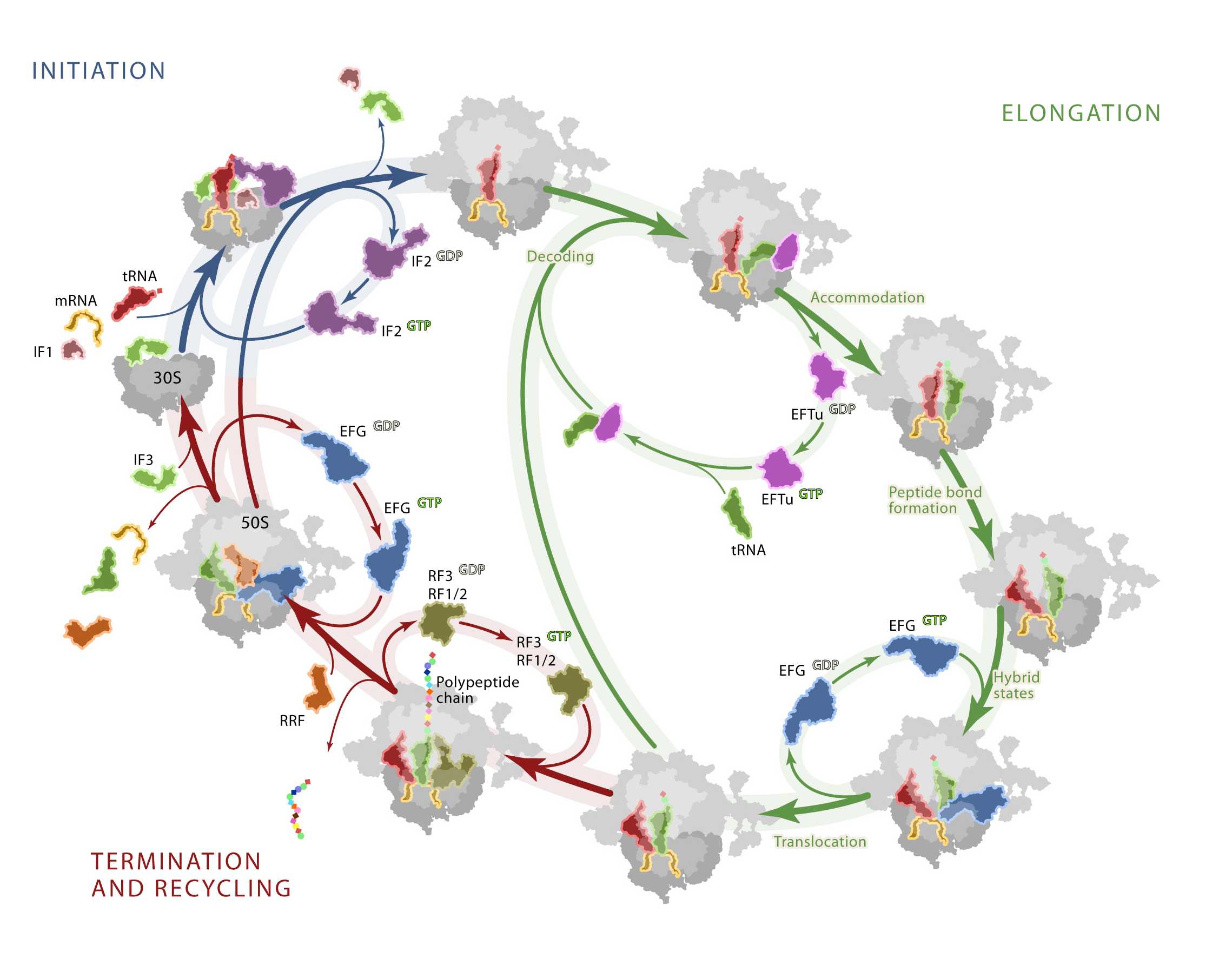
The translation machinery is responsible for the accurate conversion of the genetic information within messenger RNA (mRNA) into a corresponding polypeptide sequence. The ribosome provides the platform on which the mRNA is recognized and “decoded” by transfer RNAs (tRNAs). The process of translation occurs in four principal stages: initiation, elongation, termination and recycling. The process of initiation, facilitated by initiation factors, places the unique initiator fMet-tRNAfMet at the P site of the ribosome where it is interacts with the start codon of the mRNA. Following initiation, ribosomes enter into the elongation phase of protein synthesis. Elongation lies at the heart of protein synthesis and involves the entry and movement of tRNAs through the three tRNA binding sites (A → P → E) of the ribosome in a cyclic fashion. The number of elongation cycles is dictated by the length of the ORF and the polypeptide being synthesized. During elongation, aa-tRNAs are selected by the ribosome according to the mRNA codon presented at the A-site of the 30S subunit A site, in a process referred to as decoding. The delivery of the aa-tRNAs is a multistep, induced fit process that is facilitated by elongation factor Tu (EF-Tu) and utilizes GTP hydrolysis. Complementary base-pairing interactions between the tRNA anticodon and mRNA codon stimulates EF-Tu to hydrolyse GTP and dissociate from the ribosome, allowing aa-tRNA to fully accommodate into the PTC. Peptide-bond formation occurs between adjacently bound peptidyl- and aminoacyl-tRNAs and transfers the growing polypeptide chain from P-site tRNA to A-site tRNA, leaving deacylated-tRNA in the P site. This ribosomal state, referred to as the pre-translocation complex (PRE state), is highly dynamic in nature. In this complex, A- and P-site tRNAs reversibly oscillate between classically-defined A/A and P/P configurations and so-called hybrid states (A/P, P/E) wherein the 3’-CCA ends of both tRNAs move with respect to the large subunit while remaining relatively fixed with respect to the small subunit. The elongation cycle progresses by the translocation of A- and P-site tRNAs with respect to the small subunit. This complex multistep process, mediated by elongation factor-G (EF-G) catalyzed GTP hydrolysis, moves the mRNA-tRNA2 complex into the P- and E-sites. In so doing, a post-translocation complex (POST state) is formed, and the next downstream mRNA codon enters the A site. As repetitive elongation cycles continue, with the ribosome alternating between globally distinct PRE and POST states, the nascent polypeptide chain passes through a tunnel in the large ribosomal subunit and emerges into the cytoplasm where protein folding takes place. When a stop codon of the mRNA ORF enters into the A-site, the ribosome diverts into the termination phase. Here protein release factors specifically recognize the stop codon and mediate water-catalyzed hydrolysis of the nascent peptide from P-site tRNA. The ribosomal subunits are then separated from deacylated tRNA and mRNA through a multistep, factor-mediated process termed recycling.
Regulating the ribosome
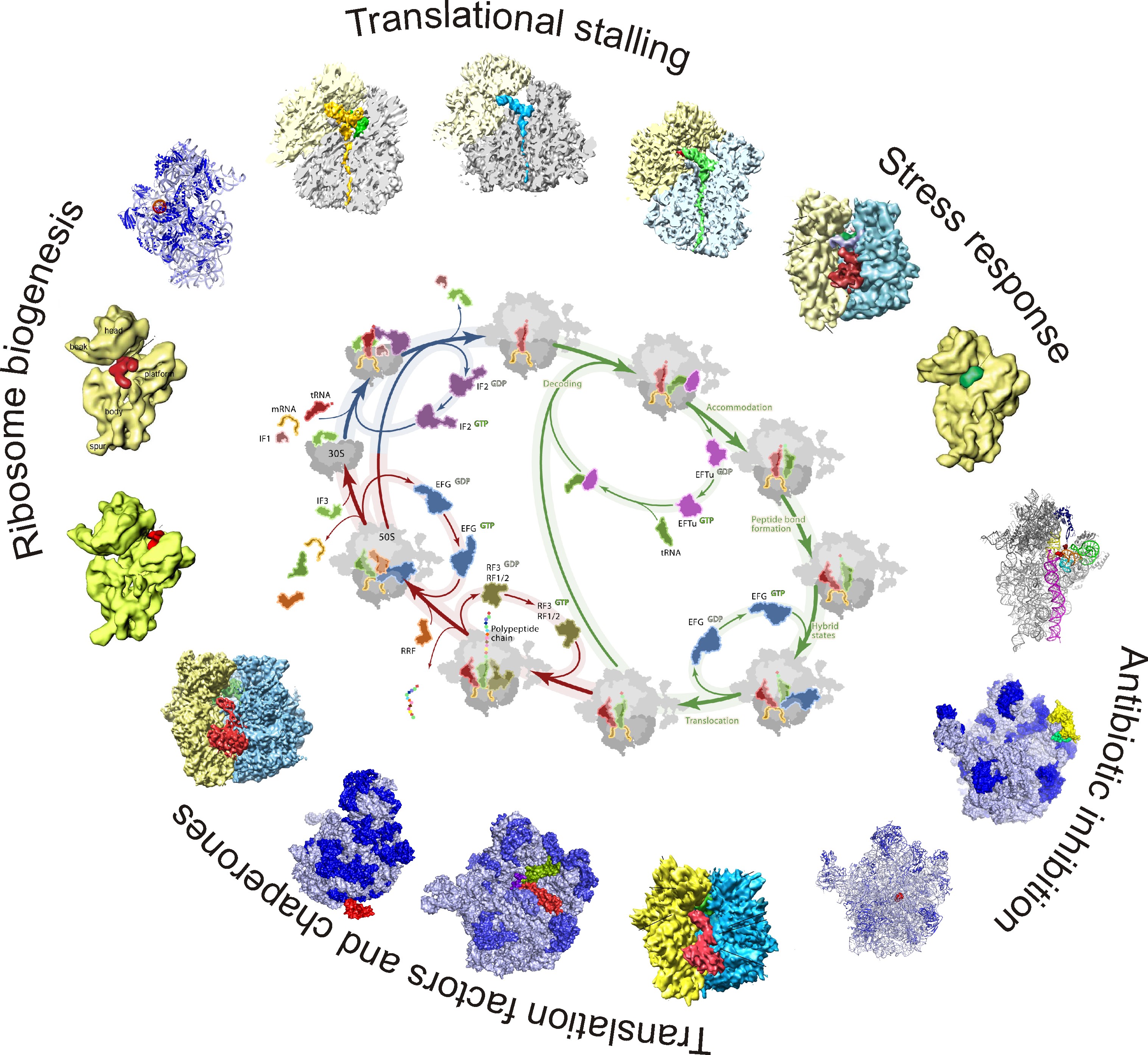
The goal of our research is to understand at a molecular level how the ribosome and protein synthesis are regulated in the cell. This is achieved by using a combination of biochemical and structural analysis (cryo-electron microscopy).
